
Mass spectrometry for quantitative proteomics in the Olsen Group
The Olsen Group address unsolved questions in cell signalling. The group aims to characterize how signalling networks regulate cellular responses differentially when mediated by phosphorylation and other modifications.
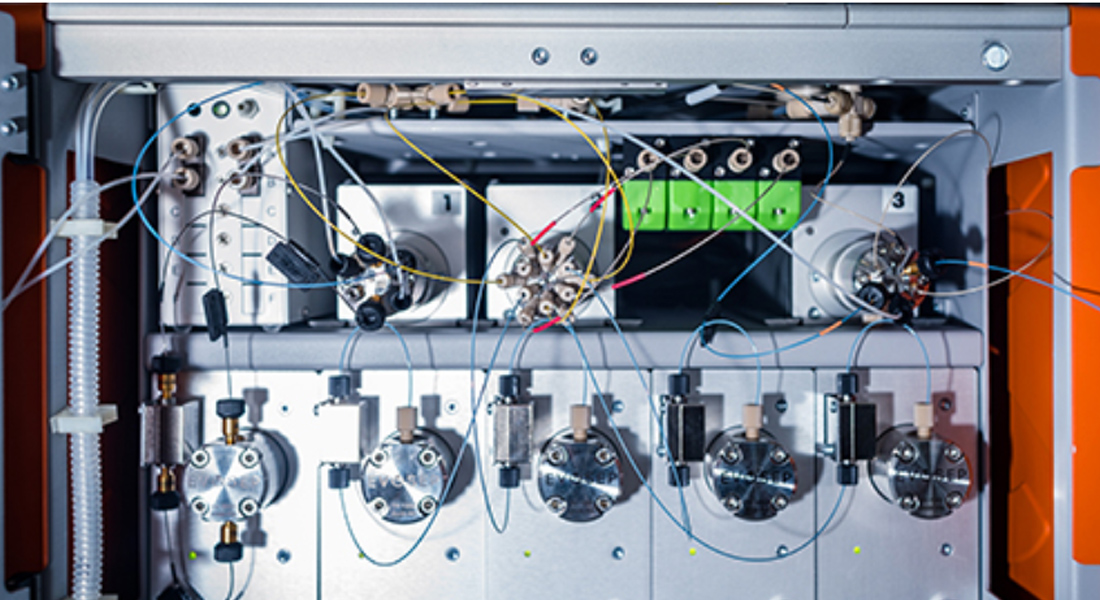
The major scientific focus area for the Olsen Group is quantitative, high-resolution mass spectrometry-based proteomics with specific emphasis on mass spectrometric technology developments and biological applications.
The group has a long-standing interest in applying their proteomics technology developments to systems-wide analyses of dynamic post-translational modifications such as site-specific phosphorylation that regulate essentially all cellular signal transduction pathways.
“We are interested in understanding how different growth factors binding to the same receptor on the outside of cells activate differential signaling pathways and signals different cellular outcomes – a phenomenon known as functional selectivity. Developing and applying quantitative mass spectrometry-based proteomics technologies allows us to generate quantitative maps of modifications, interactome and protein expression changes as a function of stimuli and stimulation time. These datasets form the basis of hypothesis generation that we test using different cell-based assays,” says Professor and Group Leader Jesper Olsen.
Offline peptide fractionation and enrichment methods combined with high-resolution orbitrap tandem mass spectrometry acquisition strategies are continuously developed by the Olsen Group with the aim to be more robust, reproducible, and rapid.
Elucidating molecular mechanisms explaining how different growth factors targeting the same receptor result in different cellular outcomes. These studies identified specific tyrosine phosphorylated residues in signaling proteins that serve as molecular switches, which determines cell fate.
Francavilla et al, Mol Cell 2013; Francavilla et al, NSMB 2016.
Benchmarking and optimizing MS acquisition strategies on Q Exactive Orbitrap instrument generations for deep human proteome analysis using shotgun proteomics.
Kelstrup et al, J Proteome Res. 2012, 2014, and 2017.
An optimized proteomics workflow that enable comprehensive analysis of human proteomes with coverage similar to next-generation RNA-seq with depth of more than 12,000 different protein-coding genes in about a day of measurements.
Bekker-Jensen et al, Cell Systems 2017.
Staff list
| Name | Title | Phone | |
|---|---|---|---|
| Agnete Witness Præst Jensen | PhD Fellow | ||
| Charlotte Hjort | PhD Student | ||
| Cristina Hernandez Rollan | Researcher | +4535337630 | |
| Jesper Velgaard Olsen | Executive Director | +4535325022 | |
| Kristina Bennet Emdal | Assistant Professor | +4535331389 | |
| Leander Roeland van der Hoeven | PhD Fellow | ||
| Maico Yannic Lechner | PhD Fellow | ||
| Ole Østergaard | Researcher | +4535326090 | |
| Tanveer Singh Batth | Research Consultant | +4535330688 | |
| Ulises Hernández Guzmán | Researcher | +4535337222 |





