Protein can release trapped histones in the cell
The heat-shock protein DNAJC9 has a surprising dual histone chaperone functionality a new study in Molecular Cell shows. The protein is the first shown to actively release histones that are trapped in non-specific interactions with DNA, directing them back to their job of organizing chromatin in the cell.

In the cell nucleus, histones play a crucial role packaging DNA into chromatin. Histones are however very sticky to both DNA and RNA, so to ensure they are transported to the cell nucleus after synthesis and bind to the right portion of DNA to organize the chromatin, they are guarded by complexes of histone chaperones. Histone chaperones are proteins that bind to histones to help protect them from non-specific binding events until they reach their goal. This process fails sometimes and histones get stuck during their supply to chromatin without any purpose.
In a study published in Molecular Cell, researchers have shown that the protein DNAJC9 holds an important role in safeguarding histones and thereby chromatin.

Histone proteins wrap DNA into structures called nucleosomes, there are around 30 million in each cell in a structure called chromatin which regulates access to the DNA code.
Active fixer joins passive bystanders
“Until now researchers have assumed that histone chaperones only act to passively shield histones. We have found out that DNAJC9 actively engages the cellular protein folding machinery – which means it actively recruits enzymes and molecular chaperones to redeploy histones that have been trapped,” says assistant professor Colin Hammond, who has lead the study. Hammond is part of the Groth Lab which is part of the Protein Memory Program in the Novo Nordisk Foundation Center for Protein Research (CPR) at University of Copenhagen.
Once released from their trapped condition histones can re-engage histone chaperones like MCM2 and be assembled into nucleosomes to organize chromatin. When the protein DNAJC9 is mutated to lose its ability to recruit the protein folding machinery, the histones stay trapped and are thereby lost for proper chromatin deposition.
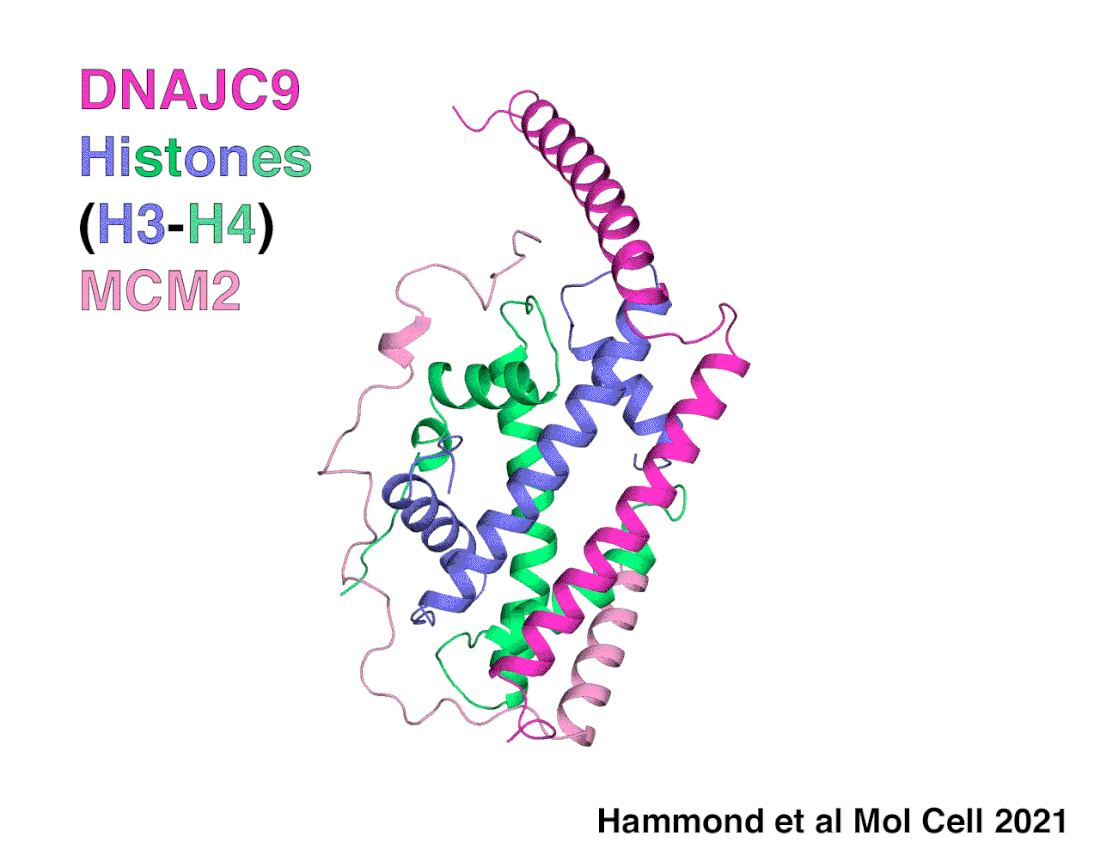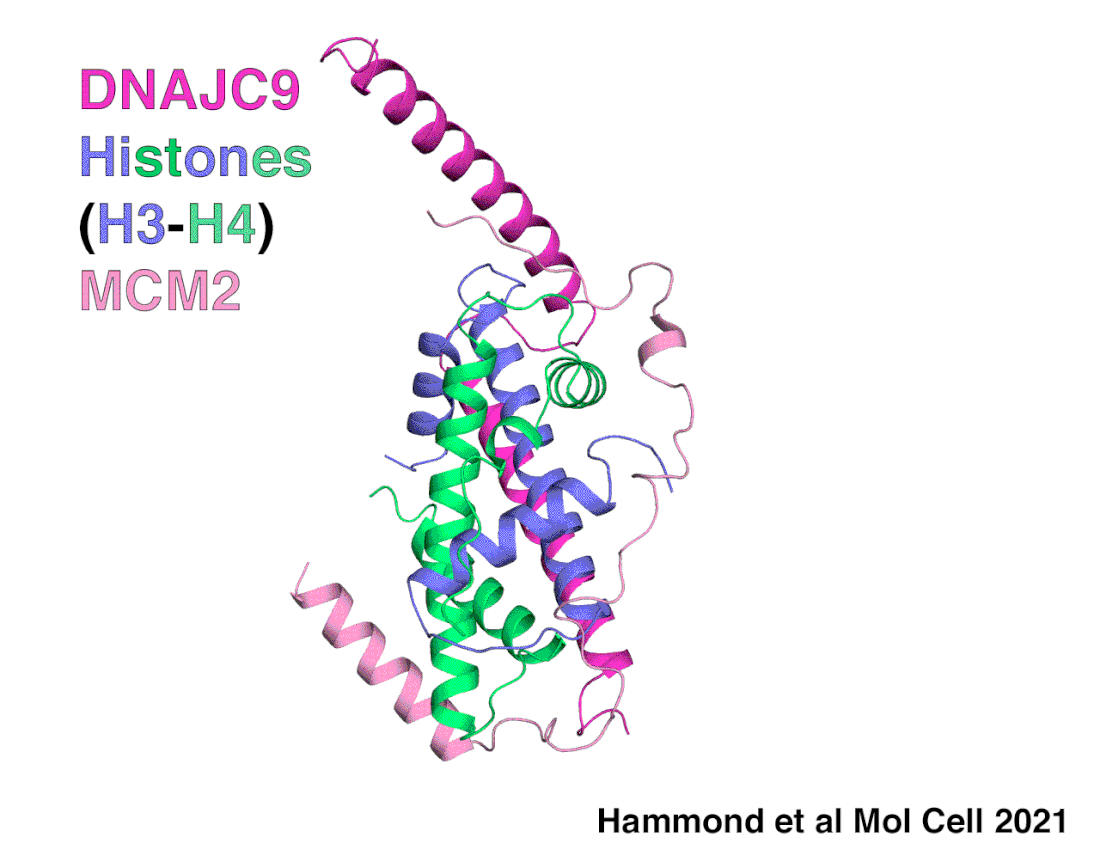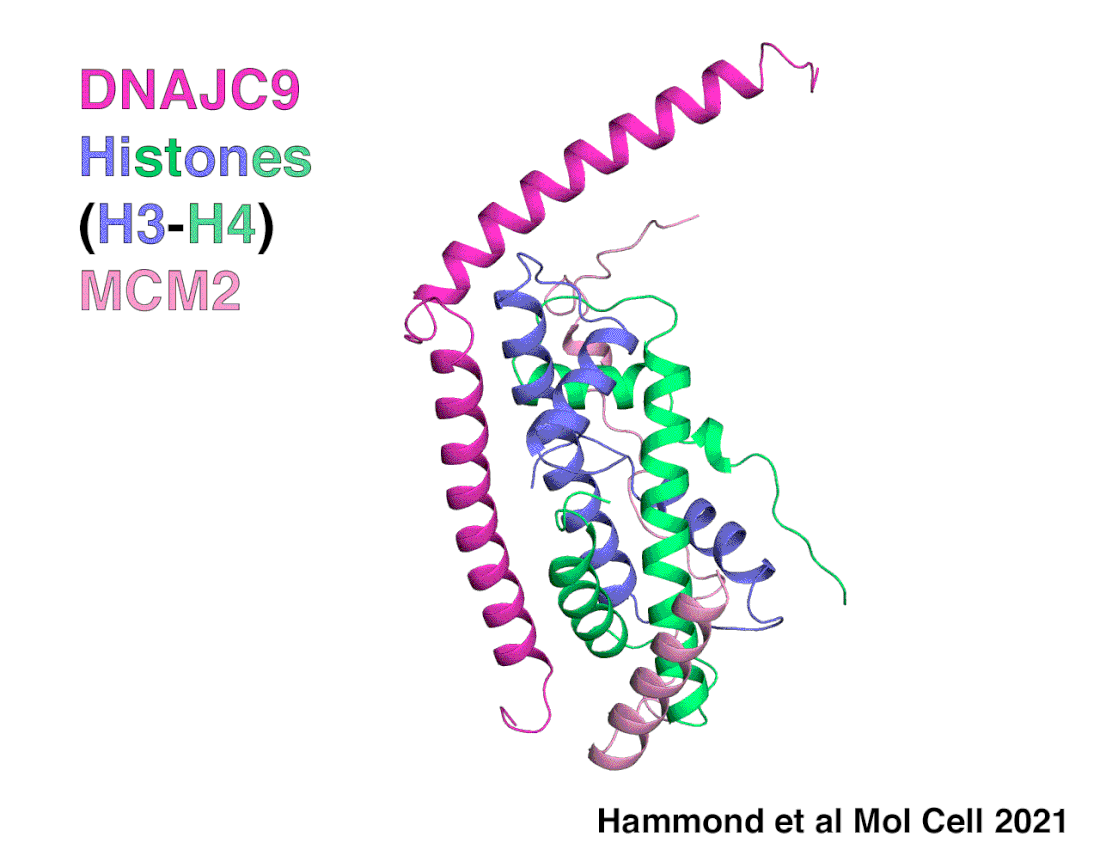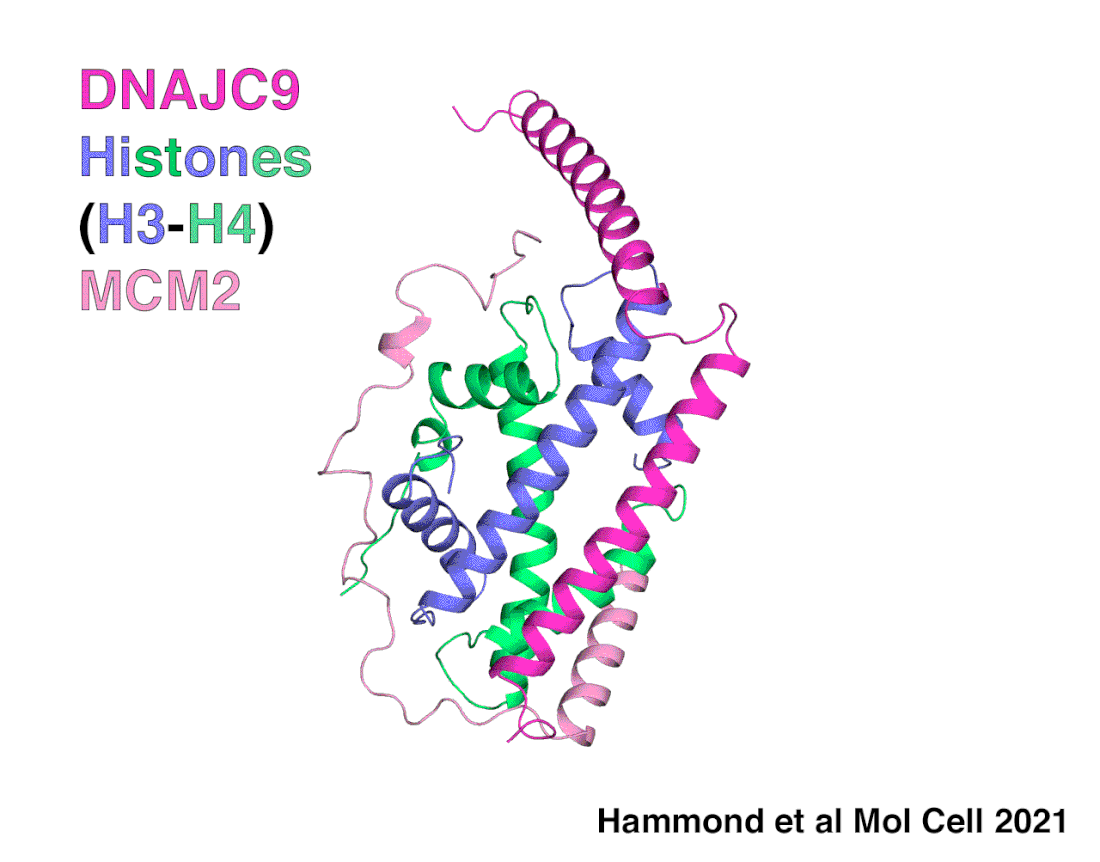
A snapshot of DNAJC9 delivering histones to the histone chaperone protein MCM2
“This means that traditional histone chaperones cannot fully protect histone proteins from spurious interactions. Rather, the cell is dependent on the combined action of molecular chaperones and histone chaperones to safeguard these fundamentally important proteins during their dynamic lives,” Colin Hammond underlines.
Strong collaboration necessary
The study is based on a strong international collaboration. Co-first author Hongyu Bao in Hongda Huang’s lab and other partners from Southern University of Science and Technology in China, and Dinshaw Patel from Memorial Sloan Kettering Cancer Center, provided the structural discoveries of DNAJC9. The functional analysis was spear-headed by Colin Hammond in close collaboration with colleagues from the Proteomics Program at CPR, Ivo Hendriks and Michael Lund Nielsen, to dissect the function of the protein through in depth proteomic assays.
“This study shows the power of combining structural biology with functional analysis, with the structural results informing the proteomic work and vice versa. We have also really benefitted from collaborating closely with our colleagues here at CPR. Being able to discuss the technical issues directly was key to the progress we made on the mass spectrometry side of the research. Combined we have essentially been able to provide a comprehensive characterization of DNAJC9 functionality,” says Colin Hammond.
Relevant insights for cancer research
DNAJC9 is an essential protein in many cancer cell types and the levels of the protein correlate with the rates at which cancer cells proliferate. Chromatin in cancer cells may be more reliant on DNAJC9 compared to regular cells, and if this is the case DNAJC9 could be a target for the development of future cancer treatments.
“Although it’s still early days, we hope this fundamental advance in our understanding of DNAJC9 biology helps to pinpoint a function essential for cancer cell viability with therapeutic potential,” Colin Hammond says.
More info:
The project originally started at BRIC and continued when the Groth Lab moved to CPR in 2020.
This research was funded in part by Anja Groth’s ERC consolidator grant.
For more details see: Hammond et al 2021 “DNAJC9 Integrates Heat Shock Molecular Chaperones into the Histone Chaperone Network” published in Molecular Cell, 14 April 2021.
