High-throughput and high-sensitivity method to analyse hundreds of cancer phosphoproteomes
A new paper from the Olsen group at CPR presents a new strategy to identify activated signaling proteins that can serve as biomarkers or drug targets in diseases. The strategy is based on mass-spectrometric analysis and will enable researchers to analyse the phosphoproteome of patient biopsies with a much higher throughput, reproducibility and sensitivity, allowing its implementation in clinical and biomedical laboratories.
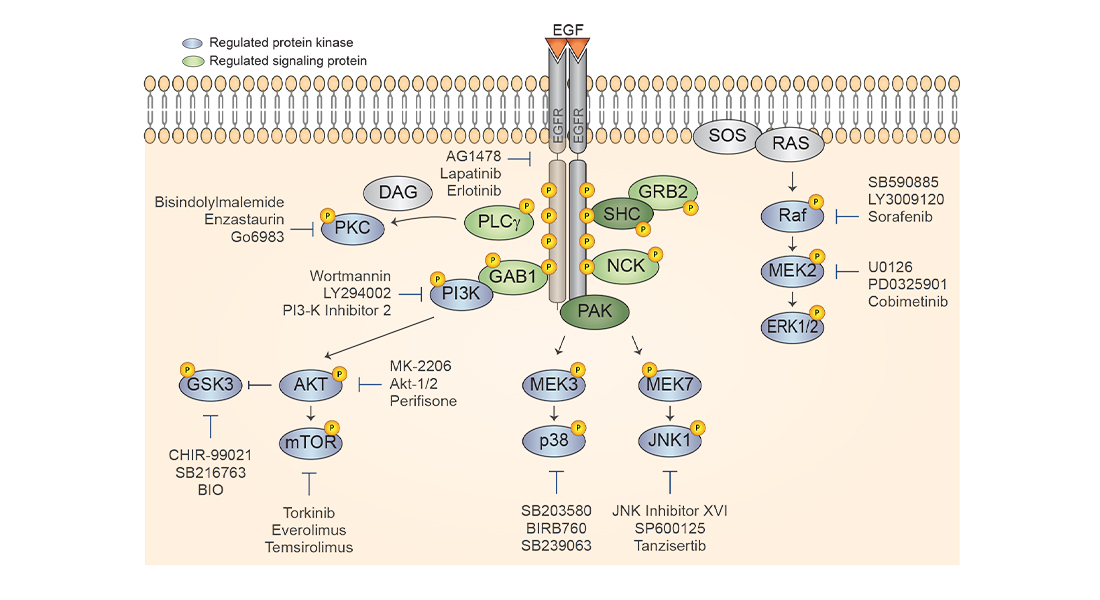
The study, which is published today in Nature Communications, is authored by members of the Olsen Group at CPR and collaborators from Biognosys AG in Switzerland and describes a new mass-spectrometric analysis strategy based on data-independent acquisition (DIA) to rapidly profile hundreds of phosphoproteomes with high quantitative accuracy and reproducibility.
Prominent drug targets
Site-specific protein phosphorylation is one of the most important post-translational modifications (PTMs), which the cell is using to modulate protein function in a rapid and reversible manner. The baseline phosphoproteome status can therefore inform about the cellular signalling networks that are activated or suppressed in a given cell state. Protein phosphorylation is catalysed by the action of an enzyme class called protein kinases and their central role in regulation of phosphoproteome status make them prominent drug targets for the pharmaceutical industry.
MSmed - developing breakthrough technology that enables patient-oriented proteome profiling
MSmed is an EU-funded FET-OPEN project that aims to make mass spectrometry so robust and powerful that it will be present in every biological laboratory and in every clinic.The acronym MSmed is short for “Mass spectrometric technology for next generation proteomics in systems medicine”. The participants of the MSmed consortium are leaders in proteomics technologies from academia and industry with out-standing track-records in advancing both instrumentation as well as its application in biological and disease contexts.
In the new study, the researchers systematically analysed the impact of 30 kinase inhibitors in different concentrations on the EGF-dependent phosphoproteome in cancer cells. This approach clearly demonstrates the power of the optimized phosphoproteomics strategy by identifying the growth factor activated kinase signalling networks in cancer cells that are perturbed by the different inhibitors (see more details about signaling via the EGF receptor in the figure).
The research project is part of MSmed, an EU-funded project that aims to make mass spectrometry, which is an advanced and highly specialised technology, accessible to biological laboratories and clinicians. So far, global phosphoproteomics has lacked the scalability necessary for high-throughput implementation in clinical and biomedical laboratories as it required milligrams of starting material and hours to days of mass spectrometric analysis time for each individual sample. Consequently, the mass spectrometric technology described in the new study has a number of potential benefits for human health in the future.
Towards phosphoproteome profiling of patients
DIA: Data-independent acquisition
DIA is a mass spectrometric acquisition strategy that systematically measures all peptide ions regardless of their intensity. DIA therefore provides better sensitivity and accuracy for quantification compared to data-dependent acquisition (DDA) shotgun proteomics, where peptides are analysed according to their abundance.In a DIA analysis, all (phospho)peptides within a mass-to-charge (m/z) window are co-fragmented and the resulting fragments analysed together. This analysis is repeated as the mass spectrometer goes through the full mass range, facilitating systematic measurement of all peptide ions regardless of their intensity. This overcomes the precursor selection problem of DDA shotgun proteomics.
In addition, the new strategy circumvents the need for elaborate data processing algorithms and software solutions normally required for DIA analysis, which makes the strategy much more accessible to non-expert laboratories.
To realise the ambitious goal of the MSmed project, the researchers developed and optimised an innovative mass spectrometric analysis strategy based on data-independent acquisition (DIA, see fact box for more information) for phosphoproteome profiling, which overcome some of the reproducibility issues associated with conventional proteomics. In addition, the collaborators from Lukas Reiter’s team at Biognosys developed and implemented a PTM localization algorithm into the software Spectronaut, the most advanced and popular software for DIA proteomics, which as a result can now also analyse DIA-based phosphoproteomics data.
In summary, the method described in the new study makes the use of mass spectrometry much more accessible to non-expert laboratories. In the near future this technology will make it possible to perform proteome profiling on patients, which will provide clinicians with specific knowledge that can support personalised treatment strategies.
The results are published in Nature Communications on Friday Feb. 7th in the study Rapid and site-specific deep phosphoproteome profiling by data-independent acquisition without the need for spectral libraries
Contact:
Professor Jesper Velgaard Olsen
jesper.olsen@cpr.ku.dk
+45 35 32 20 22
