Specificity determinants for PTM modifying enzymes in the Nilsson Lab
The goal of the Nilsson lab is to understand the basic principles of how PTM modifying enzymes achieve specificity and how these mechanisms control cellular signaling.

We are focusing on short linear motifs (SLiMs) as specificity determinants for protein phosphatases and ubiquitin ligases that are two major classes of post translational modification (PTM) regulating enzymes. Through collaborative interdisciplinary research we discover and characterize novel SLiMs hereby uncovering the mechanisms of how PTM modifying enzymes select their substrates to control biological pathways. To connect our basic discoveries to translational applications we develop small molecule inhibitors aimed at precisely disrupting SLiM based interactions.
Regulation of protein activity by PTMs is a major regulatory mechanism. Despite tremendous progress in identification of PTMs the fundamental principles regulating their addition or removal are poorly understood. An emerging concept for many PTM modifying enzymes is that SLiMs in substrates or substrate specifiers confer specificity. Identification and characterization of these SLiMs therefore provides important insight into PTM modifying enzymes.
”Our aim is to bring the study of protein phosphatases and ubiquitin ligases from the realm of passive observation to a situation where precise engineering of signaling pathways is possible,” says Professor and Group Leader Jakob Nilsson.
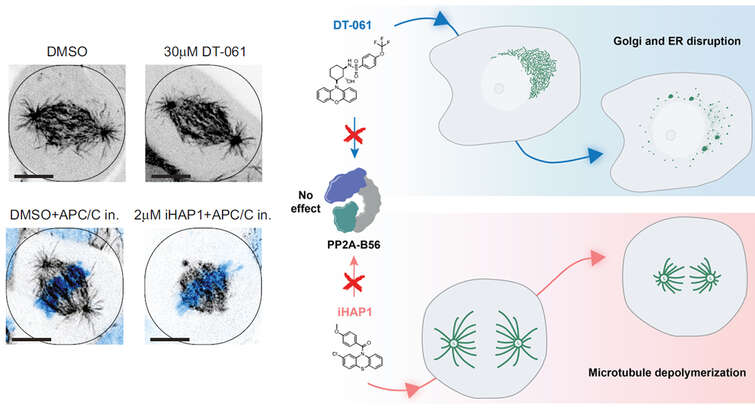
The group has uncovered general principles of substrate selection for the PP2A-B56 phosphatase (Hertz et al, Mol Cell 2016) and PP4 (Ueki et al, Mol Cell 2019). This work has shown that short linear motifs (SLiMs) are key determinants of specificity for phosphatases and that these interactions are regulated by phosphorylation of the SLiMs. A careful characterization of SLiMs allows for a system wide understanding of pathways regulated by protein phosphatases. “In addition to providing a full picture of pathways regulated by phosphatases SLiM mediated interactions are also ideal targets for small molecule inhibitors and we are actively pursuing this to develop the first class of specific inhibitors” says Jakob Nilsson.
The group has recently expanded these efforts to identify SLiMs that confer specificity to ubiquitin ligases as part of the UBIMOTIF network (https://ubimotif.ku.dk). SLiMs are often recognized by ubiquitin ligases to target a protein for degradation which is exploited in PROTAC technologies. Identifying SLiMs for ubiquitin ligases has the potential to expand the applications of PROTAC technologies.
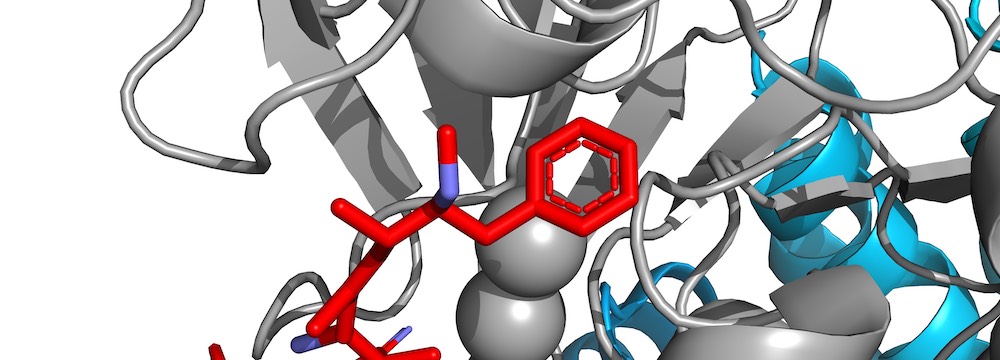
”A conserved motif provides binding specificity to the PP2A-B56 phosphatase”
We identify the LxxIxE motif as a general binding motif for PP2A-B56 and use this to dissect the function of PP2A-B56 in cytokinesis and transcriptional responses. This work is the first to describe a general principle for PP2A specificity and provides system-wide information on its function.
”Distinct kinetics of serine and threonine dephosphorylation is essential for mitosis”
We provide the first biological evidence to show that the inherent phosphothreonine preference for PP2A is important. This preference is used to control temporal timing of mitotic events.
"The Ebola virus nucleoprotein recruits the host PP2A-B56 phosphatase to activate transcriptional support activity of VP30"
We identify PP2A-B56 as a novel host factor for the Ebola virus and mechanistically dissect the role of the phosphatase in viral infection. We furthermore validate PP2A-B56 as a therapeutic target for inhibition of Ebola infection.
Staff list
| Name | Title | Phone | |
|---|---|---|---|
| Jakob Nilsson | Professor, Group Leader | +4535325053 | |
| Johanna Kliche | Postdoc | +4535324150 |