Olsen Group defines the HeLa cell proteome
Most popular and famous human cell line gets its complete proteome defined
The world’s most famous human cell line has had its complete set of expressed proteins, its proteome, determined by mass spectrometry-based proteomics (ref. Bekker-Jensen et al, Cell Systems 2017). Based on transformed epithelial cells derived from a cervical cancer tumor biopsy of a patient named Henrietta Lacks, HeLa was the first human cell line established in culture. HeLa cells are by far the most widely-used cell line in biomedical research and it contributed significantly to the development of the vaccine against the polio virus. HeLa cells were also the source for the discovery of the link between human papilloma virus (HPV) and cervical cancer for which Harald zur Hausen got the Nobel Prize in 2008. This laid the foundation for the development of the successful HPV Cancer Vaccine.
CPR researchers from Jesper Olsen’s group and collaborators from Aarhus University Hospital analyzed the HeLa proteome using a new powerful mass spectrometry-based proteomics technology. The authors established the protein copy numbers of 12,209 protein-coding genes in HeLa cells, which is comparable to the coverage of the achieved by next-generation RNA-Seq technology. The unprecedented high protein sequence coverage achieved also enabled deep identification of 18,237 unique post-translational modification sites including more than ten thousand phosphorylation sites without specific enrichment and directly established their stoichiometry in HeLa cells.
Bekker-Jensen et al. show that proteomics can now provide an essentially complete HeLa proteome. They provide measurements acquired simultaneously for more than 12,200 protein-coding genes, 10,000 phosphorylation sites, and 7,000 N-acetylation sites. Their approach is fast, accessible, and requires modest amounts of starting material, making it easily extendable to other human cell lines and patient samples.
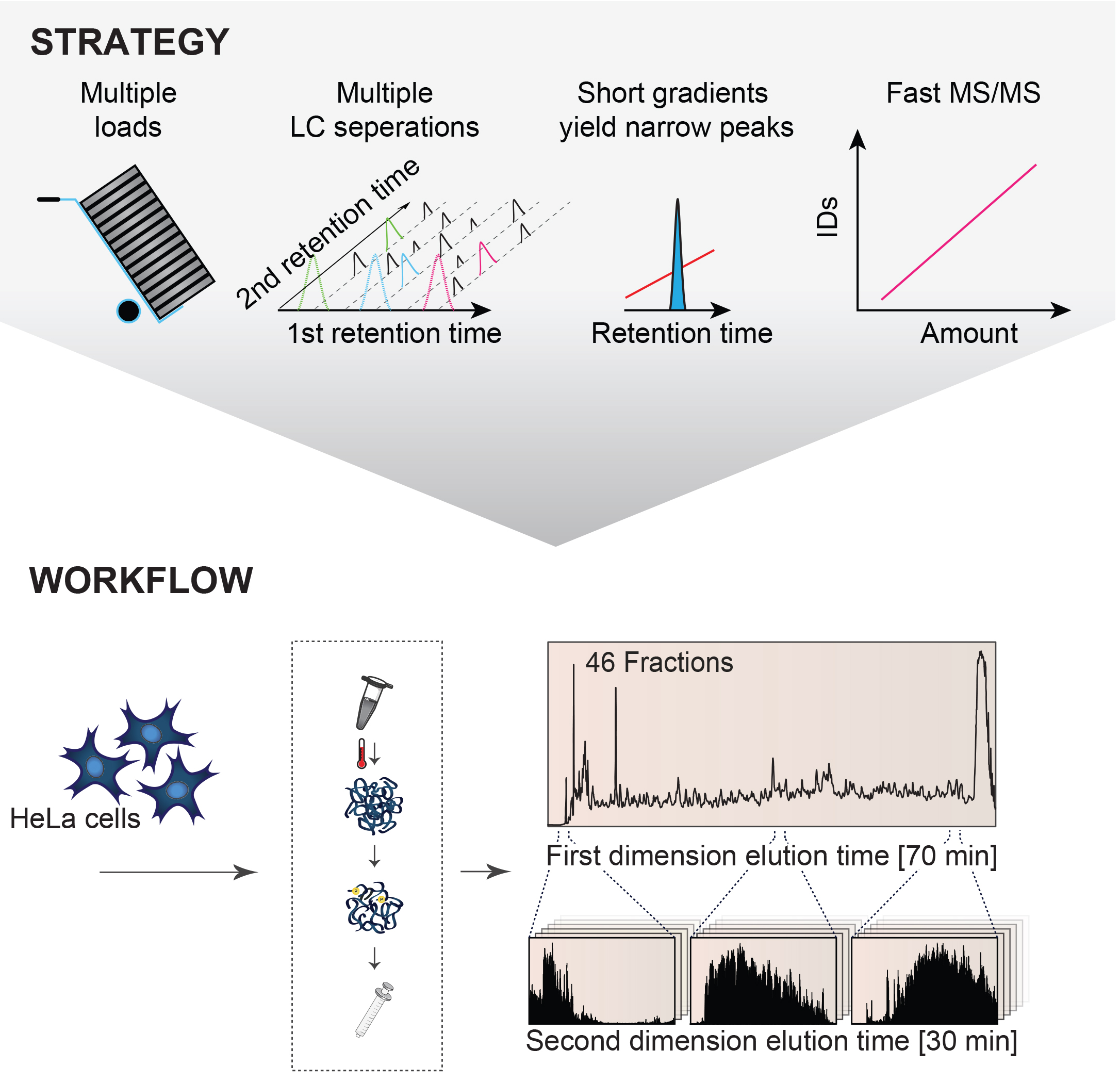
Truly comprehensive proteome analysis is the Holy Grail in proteomics for enabling routine systems biology analyses and biomarker discoveries. Despite decades of tremendous developments in the proteomics technologies, complete proteome characterization of human cells have remained a formidable task. This is mainly due to the high dynamic range of a human cell proteome spanning at least seven orders of magnitude. To overcome this challenge and routinely perform comprehensive analyses of human proteomes the CPR researchers developed so-called multi-shot proteomics, an optimized approach for comprehensive proteome analysis with high sample capacity based on offline peptide high pH reversed-phase chromatography collecting high numbers of fractions in combination with short LC-MS/MS gradients and high peptide sequencing speed. The technology has been streamlined for one day mass spectrometric measurements of human proteomes from cells and tissues. The authors proved the broad applicability of the technology by applying it to a variety of sample types including different human cancer cell lines and patient organ biopsies achieving similar comprehensive protein coverage in all cases. These experiments firmly established the capability of this proteomics technology to enable comprehensively analysis of any mammalian cell or tissue proteome in a quantitative manner with sufficient throughput for clinical applications with larger patient cohorts.
REFERENCE: http://www.cell.com/cell-systems/fulltext/S2405-4712(17)30188-6
An Optimized Shotgun Strategy for the Rapid Generation of Comprehensive Human Proteomes
Authors: Dorte B. Bekker-Jensen, Christian D. Kelstrup, Tanveer S. Batth, Sara C. Larsen, Christa Haldrup, Jesper B. Bramsen, Karina D. Sørensen, Søren Høyer, Torben F. Ørntoft, Claus L. Andersen, Michael L. Nielsen, and Jesper V. Olsen
Published Online: June 07, 2017
