Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry
 Phosphatidylinositol is a negatively charged phospholipid that represents less than 5% of the total phospholipid pool at the cytosolic side of eukaryotic cell membranes. Phosphatidylinositol can be phosphorylated by a variety of kinases on position three, four or five of the inositol ring in seven different combinations. Phosphorylated forms of phosphatidylinositols, known as phosphoinositides (PIPs), play important roles in lipid-mediated cell signaling, membrane trafficking and diseases involving these processes. PIPs can act as precursors for secondary messengers or interact directly with proteins to orchestrate spatio-temporal activation of down-stream signaling components. Despite great biological interest in the PIP signaling pathways, our knowledge of proteins that specifically interact with PIPs is still limited. This is mainly due to the absence of ‘unbiased’ technologies for detecting PIP interactions on a proteome-wide scale.
Phosphatidylinositol is a negatively charged phospholipid that represents less than 5% of the total phospholipid pool at the cytosolic side of eukaryotic cell membranes. Phosphatidylinositol can be phosphorylated by a variety of kinases on position three, four or five of the inositol ring in seven different combinations. Phosphorylated forms of phosphatidylinositols, known as phosphoinositides (PIPs), play important roles in lipid-mediated cell signaling, membrane trafficking and diseases involving these processes. PIPs can act as precursors for secondary messengers or interact directly with proteins to orchestrate spatio-temporal activation of down-stream signaling components. Despite great biological interest in the PIP signaling pathways, our knowledge of proteins that specifically interact with PIPs is still limited. This is mainly due to the absence of ‘unbiased’ technologies for detecting PIP interactions on a proteome-wide scale.
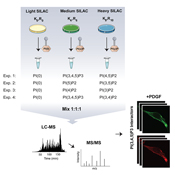
A research team led by Associate Professor Michael L. Nielsen at Novo Nordisk Foundation Center for Protein Research, in collaboration with researchers from the Max-Planck Institute in Martinsried, Germany, has utilized high-resolution quantitative mass spectrometry for mapping of phosphoinositide (PIP) interacting proteins in human cells, and profiled their binding specificities towards all seven PIP variants. The analysis revealed 405 PIP-binding proteins, more than the total number of proteins containing either phospho- or ubiquitin-binding domains.
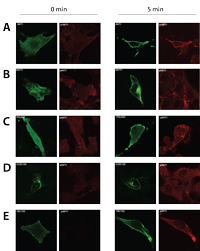
Several novel PIP-binding proteins are validated by translocation and inhibitor assays, confirming that the presented methodology targets direct interactors. The established dataset furthermore revealed a consensus motif for PI(3,4,5)P3 interacting pleckstrin homology (PH) domains, which enabled the researchers to perform in silico identification of novel phosphoinositide interactors. Moreover, the generated data revealed novel insights into known PIP-binding protein families, as demonstrated by the identification of novel PIP binding proteins and extending the known PIP binding specificity of DOCK family members. Collectively, the presented resource dataset provides a valuable resource of PIP-binding proteins in human cells.
Title: Specificity and commonality of the phosphoinositide-binding proteome analyzed by quantitative mass spectrometry
Authors: Stephanie Jungmichel, Kathrine B. Sylvestersen, Chunaram Choudhary, Steve Nguyen, and Michael L. Nielsen

