New method allows researchers to find protein imbalances causing diseases like cancer
Researchers from the University of Copenhagen have developed a first-of-its-kind method called Deep Visual Proteomics. It allows them to combine the visual features of cells with their underlying molecular profile, giving the researchers an unprecedented insight into diseases such as cancer.
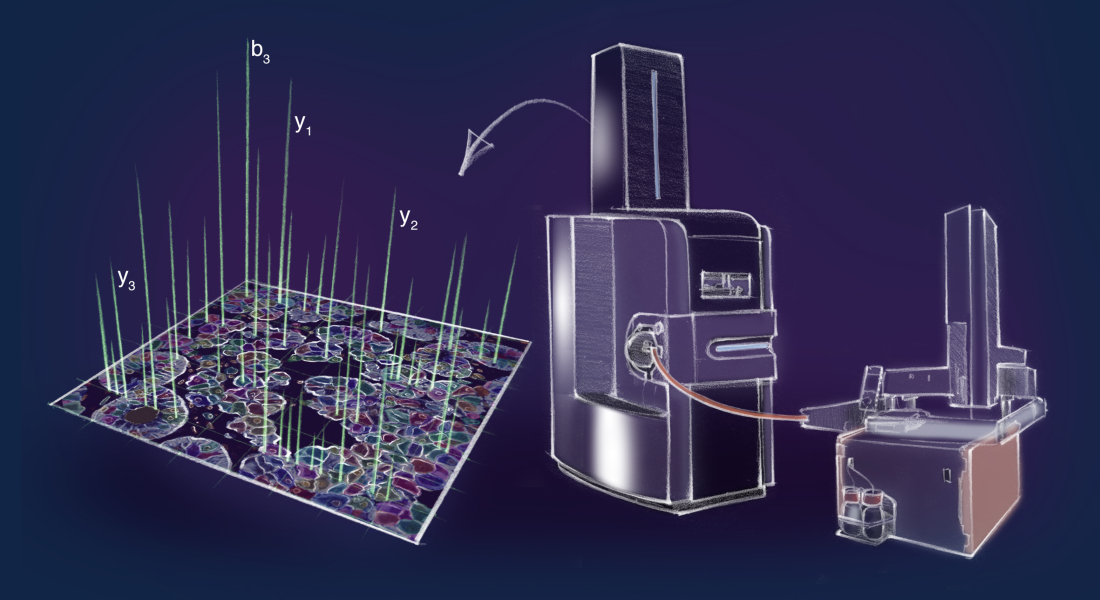
When the body gets sick, it is because of an imbalance in the smallest building blocks – the proteins. Now, researchers from the Faculty of Health and Medical Sciences at the University of Copenhagen and the Max Planck Institute of Biochemistry have developed a groundbreaking method called Deep Visual Proteomics (DVP) to help them understand what goes wrong in sick tissue like a cancer tumor.
For example, they can use DVP to find out why chemotherapy is not working or identify the protein imbalance actually causing the disease.
‘When chemotherapy does not work, it is because some of the tumor cells are chemoresistant. By dissecting a chemoresistant tumor and comparing the sick tissue with healthy tissue from the patient, we can find out which of the tumor cells are causing the resistance. Using DVP, we can then uncover the protein landscape of the sick cells to find out where the protein imbalance is. With this knowledge, we can help guide clinicians to the right treatment plan’, says Associate Professor Andreas Mund, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen.
According to the researchers, DVP can extend the possibilities of diagnosis and treatment for other diseases such as metabolic disorders and neurodegenerative diseases a well.
‘The proteins are what makes the body function – or malfunction. Diseases occur when there is a protein imbalance in the body. Our method allows us not only to uncover the protein landscape of these cells with unprecedented detail, so we can see exactly where the problem is. But also to connect the protein profile with the visual appearance of the cell. Artificial intelligence will help to detect hidden but informative patterns too subtle or complex for humans to discern by themselves and the underlying proteomic signatures identified will help to better understand the disease mechanism. It is a completely new tool that can help clinicians with diagnosis and treatment strategies’, says Andreas Mund.
A protein iceberg
Uncovering the protein landscape is very valuable because proteins are determining for health and illness and constitute drug targets. If you are sick and go see a doctor, he or she might take a blood sample. When analyzing the blood, the doctor looks for irregularities in certain proteins to find out if you have an inflammation in the body.
While the doctor might look at 10-20 proteins, DVP can analyze thousands of proteins at the same time and identify their common visual features. In this way, clinicians can connect the morphological features of the cells that are visual through the microscope with their molecular profile, giving them a much deeper insight to what is actually happening below the surface of the cells. This knowledge can be of great help when it comes to giving the right diagnosis or finding the best treatment plan.
‘You can think of it as an iceberg. The morphological features of the cell such as the shape is the visual part of the iceberg. But most of the iceberg is hidden under the surface of the water. This is the molecular profile, the protein landscape. You cannot treat a disease properly, if you only can only see the top of the iceberg. You need the whole picture’, says Andreas Mund.
Deep Visual Proteomics: A highly advanced pipeline
To prove that DVP works, the researchers have applied it to tissue samples of melanoma (skin cancer) and acinic cell carcinoma (salivary gland cancer) retrieved from the Danish biobanks.
Using artificial intelligence, DVP automatically divides the tumor cells into subgroups based on visual features such as shape, protein localization and so on. The cells are then transferred to a laser microdissection microscope, which cuts out the cells individually and shoots them into a collection well, so the different subgroups are put together. By using a completely new, ultra-high sensitivity proteomics machine, the researchers can describe the protein landscape of these subgroups with unprecedented depth, precision and accuracy.
Now, they can compare the protein landscape of the tumor cells with the protein landscape of healthy tissue from the same patient to find out where the disease causing protein imbalance is – which could then be a possible drug target.
‘This has never been done on a protein level before, and it has been possible because of the state of the art facilities and research groups working at Novo Nordisk Foundation Center for Protein Research. We have merged the best from two worlds, microscopy and proteomics, and developed a method that is a hundred times more sensitive than any previously described methods’, says Professor Matthias Mann, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen and Max Planck Institute of Biochemistry.
The new study ‘AI-driven Deep Visual Proteomics defines cell identity and heterogeneity’ is released on a preprint server in order for it to be available to everyone/the public/colleagues as fast as possible. However, it is still undergoing peer review and has not yet been published by a scientific journal.
Contact:
Associate Professor Andreas Mund
+45 35 32 50 69
andreas.mund@cpr.ku.dk
Professor Matthias Mann
matthias.mann@cpr.ku.dk
Press Officer Amanda Nybroe Rohde
+45 23 64 94 25
amanda.rohde@sund.ku.dk
